Genetics of swayback in American Saddlebred horses
Summary
Extreme lordosis, also called swayback, lowback or softback, can occur as a congenital trait or as a degenerative trait associated with ageing. In this study, the hereditary aspect of congenital swayback was investigated using whole genome association studies of 20 affected and 20 unaffected American Saddlebred (ASB) Horses for 48 165 single‐nucleotide polymorphisms (SNPs). A statistically significant association was identified on ECA20 (corrected P = 0.017) for SNP BIEC2‐532523. Of the 20 affected horses, 17 were homozygous for this SNP when compared to seven homozygotes among the unaffected horses, suggesting a major gene with a recessive mode of inheritance. The result was confirmed by testing an additional 13 affected horses and 166 unaffected horses using 35 SNPs in this region of ECA20 (corrected P = 0.036). Combined results for 33 affected horses and 287 non‐affected horses allowed identification of a region of homozygosity defined by four SNPs in the region. Based on the haplotype defined by these SNPs, 80% of the 33 affected horses were homozygous, 21% heterozygous and 9% did not possess the haplotype. Among the non‐affected horses, 15% were homozygous, 47% heterozygous and 38% did not possess the haplotype. The differences between the two groups were highly significant (P < 0.00001). The region defined by this haplotype includes 53 known and predicted genes. Exons from three candidate genes, TRERF1, RUNX2 and CNPY3 were sequenced without finding distinguishing SNPs. The mutation responsible for swayback may lie in other genes or in regulatory regions outside exons. This information can be used by breeders to reduce the occurrence of swayback among their livestock. This condition may serve as a model for investigation of congenital skeletal deformities in other species.
Introduction
Lordosis, specifically the dorsal concave curvature of the spine, is normal and healthy in most mammals. However, extreme lordosis is associated with pathology in horses (Rooney & Pickett 1967; Rooney 1969). This condition is also known in horses as swayback, lowback or softback. Rooney & Robertson (1996) noted that ‘variable degrees of lordosis seem to be common in certain lines of American Saddlebred horses’. Figure 1 shows the characteristic conformation for a horse with lordosis (Fig. 1a) and a normal horse (Fig. 1b). A familial aspect was suspected but not established in previous work. A study of swayback among Saddlebred horses led Gallagher et al. (2003) to devise a method to measure the extent of lordosis and to characterize the variation found among horses in this breed. Based on this study, a threshold for considering horses to be swayback was defined, and 5% of 294 horses were considered affected. Studies of families suggested, but did not prove, a recessive mode of inheritance.
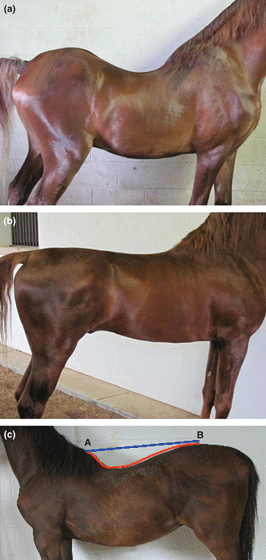
Image of a swayback 3 ‐year‐old horse (a), image of a 3 ‐year‐old horse with a normal back (b) and an image showing the method of determining the measurement of back curvature (MBC). MBC was determined as the difference between the shortest distance (blue) and the distance along the contour of the back (red) from points A and B.
In our experience, breeders are of mixed opinions regarding this trait. Swayback horses have not been routinely identified as experiencing pain, and some horses with this trait have performed well. Most breeders regard the condition as a conformation defect and avoid breeding stock with this condition. Identifying the genetic determinant(s) for this condition would provide breeders with the opportunity to better understand the genetics of the trait and use that information in their selection programs.
Traditionally, family studies have been most useful to map hereditary traits in horses (for example, Trommershausen‐Smith 1978; Bailey et al. 1997; Locke et al. 2001). However, while swayback horses are not uncommon, breeders avoid matings they believe likely to produce affected horses. Consequently, family studies are difficult. However, with the advent of the horse genome sequence and the availability of dense arrays of single‐nucleotide polymorphisms (SNPs) for whole genome association (WGA) studies, population studies can be used to investigate the genetics of traits in horses (Wade et al. 2009). The purpose of this study was to determine whether a hereditary component contributes to the swayback trait in American Saddlebred (ASB) horses and, if so, to identify the location of genes for this trait.
Materials and methods
Phenotypic assessment of lordosis
Phenotypic assessment of lordosis was based on the measurement of back contour (MBC) (Gallagher et al. 2003). Two points were chosen on the horse’s back, one at the top of the withers (the highest point of the dorsal spinous process in the region of thoracic vertebrae T2‐T3) and one on the point of the rump (the highest point on top of the horse’s hips). The shortest distance between those points was measured in centimetres and designated ‘A’. Next, the distance along the contour of the back was measured in centimetres and designated ‘B’. MBC was designated as the difference between ‘A’ and ‘B’. Figure 1c illustrates the points of measurement on a swayback horse.
Horses measured for MBC
Measurements and tissue (hair or blood) samples for ASB horses used in this study came from a sample set of 749 ASB horses collected from private and commercial farms in and around Kentucky. The average age was 7.1 years and ranged from 1 month to 29 years old. ASB horses used in the Illumina and Sequenom assays were selected from among this group of samples.
Horses for illumina assay
For the genome scan using the Illumina Equine SNP50 Chip, 40 ASB horses were selected, based on their lordosis phenotype. Twenty horses were selected based on having an MBC >8.0 cm, and 20 were selected based on having MBC <5.0 cm. The control group included 19 half‐siblings for horses in the affected group to reduce the chance of population substructure producing spurious associations.
Horses for sequenom assay
A total of 426 horses were selected for screening with 35 SNPs located in the genomic region suggested by the WGA study. Three horses were tested in duplicate to control for the quality of the testing. Twilight, the Thoroughbred horse used for the whole genome sequencing, was also tested for quality control of known SNPs located in the genomic region suggested by the WGA study. Among the ASB horses, 33 (including the 20 original cases) had values for MBC of 7.0 or greater, 287 had MBC <7.0 cm and 106 were parents siblings or offspring of swayback horses. The relatives were tested to assist with haplotype determination.
DNA extraction
DNA was extracted from blood or hair follicles for testing. DNA from blood samples was extracted using Puregene whole blood extraction kit (Gentra Systems Inc.) according to its published protocol. Hair samples were processed using 20–30 hair bulbs according to the modified protocol using a Gentra DNA purification kit. The hair bulbs were placed in 200 μl Gentra Cell Lysis solution containing 0.01 mg proteinase K (Sigma‐Aldrich) and incubated at 5 °C overnight. DNA was then purified by following remaining steps of the protocol.
Illumina equine SNP50 genotyping
Initial SNP genotyping of 40 samples in the case/control group was performed utilizing the Illumina Equine 50 SNP chip for a WGA study. DNA was provided to the core facility at the Mayo Clinic in Rochester, MN for genotyping.
Sequenom SNP genotyping
Ten SNPs from the WGA study and 39 additional SNPs from the EquCab2.0 SNP database (http://www.broadinstitute.org/ftp/distribution/horse_snp_release/v2/) were selected for testing using the MassArray iPLEX Gold assay on the Sequenom platform. Genotyping was performed at Proactive Genomics, LLC. The 49 SNPS selected were identified as being between positions 41 530 793 and 44 585 118 and are listed in Table 2. Following testing, 43 of 49 the SNPs provided quality genotyping data. Of these, 35 SNPS with minor allele frequency >0.01 were used for final association and haplotype analysis. Furthermore, 96% of the submitted samples had fewer than 0.05 genotypes missing and were thus included in analysis.
| SNP‐ID | BP | New horses | Combined horses | ||
|---|---|---|---|---|---|
| CHISQ1 | P1 | CHISQ2 | P2 | ||
| BIEC2_532523* | 41530793 | 4.416 | 0.0356 | 23.72 | 1.12E‐06 |
| BIEC2_532534* | 41604741 | 11.19 | 0.00082 | 43.72 | 3.79E‐11 |
| BIEC2_559789 | 41772507 | 0.7726 | 0.3794 | 5.543 | 0.01855 |
| BIEC2_559792 | 41823842 | 0.2935 | 0.588 | 9.102 | 0.002553 |
| BIEC2_532578* | 42062440 | 5.605 | 0.01791 | 42.03 | 8.99E‐11 |
| BIEC2_559853 | 42305090 | 0.8888 | 0.3458 | 1.363 | 0.2431 |
| BIEC2_559859 | 42395887 | 1.484 | 0.2231 | 1.669 | 0.1964 |
| BIEC2_559873 | 42482150 | 2.066 | 0.1506 | 7.259 | 0.007055 |
| BIEC2_532658* | 42603867 | 8.571 | 0.003415 | 29.79 | 4.80E‐08 |
| BIEC2_559931 | 42679616 | 0.5499 | 0.4584 | 4.836 | 0.02787 |
| BIEC2_559976 | 42757527 | 0.4859 | 0.4858 | 2.909 | 0.08809 |
| BIEC2_560005 | 42806550 | 1.517 | 0.2181 | 3.629 | 0.05679 |
| BIEC2_560049 | 42906719 | 3.046 | 0.08091 | 0.002347 | 0.9614 |
| BIEC2_532826 | 42983718 | 1.666 | 0.1969 | 13.07 | 0.0003 |
| BIEC2_560102 | 43022942 | 0.8888 | 0.3458 | 1.828 | 0.1763 |
| BIEC2_560144 | 43069858 | 0.8056 | 0.3694 | 1.711 | 0.1908 |
| BIEC2_560178 | 43195488 | 2.306 | 0.1288 | 10.9 | 0.000963 |
| BIEC2_560197 | 43241900 | 1.747 | 0.1862 | 3.929 | 0.04746 |
| BIEC2_560209 | 43319368 | 9.93 | 0.001626 | 20.93 | 4.76E‐06 |
| BIEC2_560255 | 43433802 | 1.213 | 0.2707 | 9.956 | 0.001603 |
| BIEC2_533020 | 43520722 | 6.459 | 0.01104 | 24.51 | 7.38E‐07 |
| BIEC2_560328 | 43548897 | 1.173 | 0.2788 | 10.58 | 0.001144 |
| BIEC2_560360 | 43605858 | 1.11 | 0.2921 | 9.664 | 0.001879 |
| BIEC2_560386 | 43690935 | 2.254 | 0.1333 | 13.48 | 0.000241 |
| BIEC2_560457 | 43914236 | 1.056 | 0.304 | 2.418 | 0.1199 |
| BIEC2_560475 | 43971596 | 0.9724 | 0.3241 | 2.181 | 0.1397 |
| BIEC2_560489 | 44034799 | 2.722 | 0.09899 | 11.67 | 0.000634 |
| BIEC2_533265 | 44090171 | 7.979 | 0.004733 | 26.6 | 2.50E‐07 |
| BIEC2_560585 | 44148267 | 1.407 | 0.2356 | 0.964 | 0.3262 |
| BIEC2_560618 | 44188313 | 0.09612 | 0.7565 | 1.771 | 0.1832 |
| BIEC2_533376 | 44241203 | 3.691 | 0.05472 | 19.71 | 9.03E‐06 |
| BIEC2_560705 | 44337671 | 1.659 | 0.1977 | 1.456 | 0.2275 |
| BIEC2_560742 | 44403289 | 1.312 | 0.2521 | 0.8528 | 0.3558 |
| BIEC2_560796 | 44484324 | 1.056 | 0.3042 | 8.726 | 0.003137 |
| BIEC2_533588 | 44585118 | 0.5591 | 0.4546 | 1.017 | 0.3132 |
- SNPs used for the definition of haplotypes for this region are denoted with ‘*’.
Analyses
Genotyping data analysis was performed with PLINK v1.06 (Purcell et al. 2007). Association analysis by case/control chi‐square was performed on the Illumina data. To minimize error because of the multiplicity of SNPs tested, an MPERM analysis with 10 000 permutations was performed and a statistic, EMP2 (referred to here as corrected P), verified possible associations. Once association was identified, different sized haplotypes were tested by chi‐square analysis to identify the size and location of the highest associated haplotype in the region. Selected haplotypes were phased for all horses in this study to clarify familial patterns of transmission. Chi‐square association analysis was also performed on the Sequenom data to verify association of the SNPs included from Illumina assay and to identify any new associations. The 35 SNP haplotypes were then phased to verify familial patterns and to identify possible recombination locations. Haplotype analysis was also performed in Haploview (Barrett et al. 2005) for validation of PLINK analysis. Haplotypes were identified using the HAP and PHASE options of PLINK. Haplotype frequencies were determined by direct counting.
Sequencing
To investigate candidate genes, exon sequences of two affected horses were compared to those of two unaffected horses. The two affected horses were selected as unrelated horses with MBC >7.0 and homozygous for the swayback‐associated ECA20 haplotype. Unaffected horses were selected as unrelated horses with MBC <7.0 and not possessing the ECA20 haplotype associated with swayback. When SNPS were identified and confirmed on these four horses, additional control and case horses were tested to determine whether there was an association with the swayback trait. PCR template for sequencing was amplified in 20 μl PCRs using 1× PCR buffer with 2.0 mm MgCl2, 200 μm of each dNTP, 1 μl genomic DNA from hair lysate, 0.2 U FastStart Taq DNA polymerase (Perkin Elmer, Waltham, Mass.) and 50 nm of each primer. Template product was quantified on a 1% agarose gel, then amplified with BigDye Terminator v1.1 cycle sequencing kit according to manufacturer’s instructions (Applied Biosystems), cleaned using Centri‐Sep columns (Princeton Separations Inc.), and run on an ABI 310 genetic analyzer (Applied Biosystems). Primers were designed in Primer 3 (Steve and Skaletsky 1998) using 8 intronic sequences and seven exonic sequences of TRERF1 (transcriptional regulating factor for CYP11A1), 15 exonic sequences for RUNX2 (transcription factor associate with osteoblast differentiation) and six exonic sequences for CNPY3 (regulates cell surface expression of Toll Receptor 4) (See Table S1).
Results
Distribution of MBC
The distribution of MBC values among the 749 horses in this study was similar to that found by Gallagher et al. (2003). MBC for the 749 ASB horses ranged from 0 to 17 cm. Most MBC values appeared to fall within a normal population distribution with a mean of 3.6 ± 1.9 cm, median of 4 cm and mode of 4 cm. An MBC of 7 cm or greater was selected to classify horses as affected based on this value being approximately two standard deviations from the mean.
Whole genome scan with illumina equine SNP50 beadchip
The Illumina Equine SNP50 chip (Illumina) was effective in typing DNA from the initial 40 horses. The 40 individuals had an average call rate of 0.96. After filtering for minimum minor allele frequency and genotyping, 48 165 SNPs were retained for data analysis.
Table 1 shows the top ten statistically significant results from a 2 × 2 chi‐square analysis comparing the distribution of SNPs in the DNA of affected and non‐affected horses. The most significant association was found on ECA20 for SNP BIEC2‐532523 (P = 6.69E‐06). Of the ten highest P‐values, five occurred for SNPs on ECA20 in the 531‐ kb region between positions 42 062 440 and 41 530 973. The multiplicity of comparisons can result in the spurious discovery of high chi‐square values; therefore, to control for multiple comparisons, PLINK was used to conduct a Monte Carlo simulation with 10 000 permutations to calculate a corrected P‐value (EMP2). Only the association with SNP BIEC2‐532523 on ECA20 remained significant (corrected P = 0.017).
| CHR | BP | SNP‐ID | CHISQ | P | corrected P |
|---|---|---|---|---|---|
| 20 | 41530793 | BIEC2‐532523 | 20.28 | 6.69E‐06 | 0.01699 |
| X | 51065036 | BIEC2‐1124071 | 15.66 | 7.60E‐05 | 0.2554 |
| 20 | 42062440 | BIEC2‐532578 | 15.53 | 8.12E‐05 | 0.2904 |
| 7 | 7444796 | BIEC2‐978005 | 13.73 | 0.000211 | 0.6762 |
| X | 51061227 | BIEC2‐1124068 | 13.47 | 0.000243 | 0.7091 |
| 7 | 18839866 | BIEC2‐984003 | 13.33 | 0.000261 | 0.7326 |
| 28 | 35119684 | BIEC2‐739844 | 13.33 | 0.000261 | 0.7326 |
| 20 | 41576546 | BIEC2‐532530 | 13.07 | 0.000301 | 0.7931 |
| 20 | 41604741 | BIEC2‐532534 | 13.07 | 0.000301 | 0.7931 |
| 20 | 41625809 | BIEC2‐532535 | 13.07 | 0.000301 | 0.7931 |
Among the affected horses with MBC 7 cm or greater, 17 of 20 were homozygous for the T allele of the BIEC2‐532523 T/C SNP. Among the 20 controls, only seven were homozygous for this allele. This SNP fell within a larger region of homozygosity spanning approximately 3 Mb (base position 41 604 741–44 512 270), which was identified with the homozygosity function in PLINK by scanning sliding windows of 35 SNPS, moving one SNP at a time and allowing one heterozygote per window to be considered homozygous (data not shown).
Sequenom
To verify the statistical associations found with the Illumina assay, SNPs from the candidate region on ECA20 were tested, including ten SNPS from the Illumina Equine SNP50 chip and 25 additional SNPs from the EquCab2.0 SNP database. Association chi‐squared analyses were performed separately for the additional 13 affected and 181 unaffected horses not previously included in the WGA assay. The association analyses for just these new samples are shown in Table 2 (CHISQ1 and P1). Based on these 13 affected horses, the association with BIEC2‐532523 remained statistically significant with a P‐value of 0.036. Because this was a comparison with new samples dictated by the original Illumina assay, no statistical correction is necessary to correct for multiplicity of testing, as performed for the previous experiment. Of the 35 SNPs, seven showed statistically significant associations (P < 0.05). When all 33 affected horses and 287 controls were compared, 21 of the 35 selected SNPs showed statistical significance in their distributions between the two groups based on their relative frequencies.
Haplotype analysis
Haplotypes from the region defined in the Sequenom assay (41 530 793 and 44 585 118) were compared among affected and unaffected horses. A minimum haplotype that included the maximum number of affected horses and the lowest number of non‐affected horses was identified using the four SNPs BIEC2‐532523, 532534, 532578 and 532658 and spanned 1 073 074 bp. Intervening SNPs did not affect haplotype assignment. These four SNPs allowed identification of 13 haplotypes. The haplotypes and their frequencies among the affected and non‐affected horses are shown in Table 3.
| Haplotype | Affected (N = 33) | Non‐affected(N = 287) |
|---|---|---|
| TGTG | 0.80 | 0.388 |
| CACT | 0.03 | 0.232 |
| CACG | 0.06 | 0.167 |
| TACT | 0.04 | 0.164 |
| TGCG | 0.05 | 0.019 |
| TGTT | 0.00 | 0.012 |
| CATG | 0.02 | 0.003 |
| TGCT | 0.00 | 0.003 |
| CGCT | 0.00 | 0.003 |
| CATT | 0.00 | 0.002 |
| TACG | 0.00 | 0.002 |
| TATG | 0.00 | 0.002 |
| CGTG | 0.00 | 0.002 |
- ASB, American Saddlebred.
Only four of the haplotypes had frequencies above 0.05 and had a cumulative frequency of 0.95 among unaffected horses and 0.93 among affected horses. The most common haplotype, TGTG, was associated with swayback. Haploytpe TGTG had a frequency of 0.80 among swayback ASB horses and a frequency of 0.39 among non‐affected horses. Zygosity for this haplotype is shown for affected and non‐affected horses in Table 4. Among the affected horses, 23 (70%) were homozygous for haplotype TGTG, while only 15% of the non‐affected horses were homozygous. Among the 33 swayback horses, seven (21%) were heterozygous for haplotype TGTG and three (9%) did not possess haplotype TGTG. Among non‐affected horses, 48% were heterozygous and 35% did not possess haplotype TGTG. Statistical comparison of the combined data set to the subset of horses with swayback was highly significant (P < 0.00001).
| Classification | Number of horses | TGTG homozygotes | TGTG heterozygotes | No TGTG |
|---|---|---|---|---|
| Swayback | 33 | 23 (70%) | 7 (21%) | 3 (9%) |
| Non‐affected | 287 | 44 (15%) | 135 (47%) | 108 (38%) |
| Combined | 320 | 67 (21%) | 142 (44%) | 111 (35%) |
- *Chi‐square for swayback vs. combined = 47.08, P < 0.00001.
Candidate genes
The SNPs defined an overall region of homozygosity for swayback horses ranging from 41.5 to 44.5 Mb on ECA20. The annotated horse genome at ENSEMBL genome browser (Hubbard et al. 2009) showed that this region contains 53 known and predicted genes. Three genes were selected as possible candidates based on predicted or known function in other species; sequence comparisons for TRERF1 (15 exons), RUNX2 (seven exons, eight introns) and CNPY3 (six exons) between normal and swayback horses did not identify SNPs associated with swayback. Data on SNPs found and their occurrence among the case and control horses are shown in Table S2.
Discussion
The distribution of MBC measurements in this study confirmed the results from the earlier study by describing a normal distribution of the MBC phenotype, with 5% falling two standard deviations above the mean (Gallagher et al. 2003). In that previous study, the mean was 4.05 cM, while the mean found in this study was smaller, 3.6 cM. Differences in age of horses may account for this difference, because the previous study showed a positive correlation for age and MBC; the mean age in the first study was 7.8 years, and the mean age in this study was 7.1 years.
The WGA study demonstrated the presence of a recessive gene responsible for the swayback trait in horses. This was suggested in the initial WGA study with 40 horses and the Illumina SNP chip (corrected P = 0.017). The association was confirmed in a subsequent study with a second set of affected horses using SNPs from this targeted area (P = 0.036). Using data from all horses, a haplotype showing the strongest association with swayback was identified based on four SNPs (TGTG) described in Table 3. This haplotype spanned 1 073 074 bases and the region harboured 53 known and predicted genes.
The TGTG haplotype was the most common and suggests that the haplotype occurred in the breed before the mutation causing swayback. If a gene present in this haplotype was mutated to cause swayback, then only knowing the specific mutation would allow us to distinguish between these haplotypes. Of course, mutations which subsequently occurred within the swayback‐causing haplotype might allow us to use tests for other markers to identify haplotypes completely associated with swayback, although not all horses with this gene for swayback.
The high frequency of this haplotype and this phenotype among ASB horses might be the consequence of selection by breeders. If a single copy of the gene produced a desirable phenotypic effect, such as improved gait, selection for that trait may negate selection against swayback and result in a net increase in the frequency of the gene in the breed. Comparisons of gene frequencies for the TGTG haplotype among horses with different performance phenotypes are needed to answer this question.
While the high homozygosity of this haplotype among the swayback horses demonstrated the presence of a recessive gene for the trait, not all swayback horses were homozygous for the region. We found five different haplotypes among the swayback horses, and 30% were not homozygous for the haplotype associated with the recessive swayback condition. The swayback phenotype may have multiple possible causes, of which the hereditary recessive condition is only one. While 70% of the swayback horses were homozygous for this haplotype, we observed seven (21%) swayback horses that were heterozygous and three (9%) that did not have the haplotype at all (Table 4). There may be multiple causes of swayback among Saddlebred horses, and the recessive gene suggested by this study may be only one of them. Other genes, accidents affecting skeletal integrity or even management practices may cause swayback in the absence of the recessive gene implicated by this study. Nevertheless, considering the high prevalence of this haplotype among affected horses, this hereditary recessive condition is probably the most common cause of swayback among Saddlebred horses.
As noted above, the region contains 53 known or predicted genes for the horse. We selected three genes for DNA sequencing in the hope of identifying the causative mutation. One gene in this region, RUNX2, had been implicated in skeletal defects based on information from OMIM. RUNX2 has been found to be a scaffold for factors involved in skeletal gene expression (Stein et al. 2004), and it plays a role in osteoblast and chondrocyte differentiation and migration (Fujita et al. 2004). From assessment of other likely gene functions, the candidate gene TRERF1 was identified based on its function as a transcription factor. CNPY3, a trinucleotide repeat‐containing gene, was also considered, because repeat expansion has been shown to play a role in various diseases. Such a repeat expansion within SCA1 on human 6p is responsible for spinocerebellar ataxia type 1 (Kameya et al. 1994). However, exon sequencing of the three candidate genes did not identify SNPs or other genetic markers associated with the trait.
Rooney & Robertson (1996) distinguished between senile lordosis and congenital lordosis in horses. Senile lordosis was a consequence of ageing. Congenital lordosis occurred as a consequence of hypoplasia of the articular facets of thoracic vertebrae and followed birth as a result of weight bearing (Rooney & Pickett 1967). We believe that the condition we have been investigating in ASB horses is the congenital form, because most of the affected horses were under the age of 10. However, this should be confirmed by sequential measurement of MBC in horses of different ages, concentrating especially on young horses, to determine the progression of lordosis.
Discovery of the mutation responsible for swayback is a goal that remains ahead of us. In connection with this project, exons of several candidate genes were sequenced. However, the cause of the trait could be because of aspects of gene expression which are not encoded in exons. As we learn more about the genome of animals, we realize that even the introns and the DNA between genes can play a role in gene regulation. The region of interest might be reduced by further studies using additional genetic markers from this region, including more SNPs, microsatellites or other genetic polymorphisms. Another approach to understanding this condition may be to investigate differences in gene expression between affected and unaffected horses. Discovery of a gene which shows differential expression would help to focus this work. However, the choice of tissues and the age at which horses are tested may be factors which confound such an approach.
More insight into the variation responsible for early‐onset extreme lordosis in horses may be beneficial for studies of human juvenile kyphosis and juvenile idiopathic scoliosis (IS). As with the horse, these two congenital conditions exhibit an early age of onset. Familial IS only accounts for 10% of all cases in humans, while 90% appear to be sporadic with unknown or environmental aetiological factors (Cheng et al. 2007). Through familial linkage analysis, candidate regions for IS susceptibility have been identified on human chromosomes 6p, 10q and 18q (Wise et al. 2000). In a more recent study, regions on 6, 9, 16 and 17 were identified through genome‐wide screening (Miller et al. 2005). It is of particular interest to note that the segment on HSA6 implicated in that study is syntenic with the region on ECA20 that was found in this study to be associated with swayback.
Acknowledgements
The authors are grateful to the Morris Animal Foundation and to the American Saddlebred Horse Association for funding the project. Samples were generously provided by private horse breeders. This work was in connection with the doctoral research of DC. The work is also in connection with a project of the University of Kentucky Agricultural Experiment Station as paper number 10‐00‐000.
Conflicts of interest
The authors have declared no potential conflicts.
References
Citing Literature
Number of times cited according to CrossRef: 16
- Danielle Bisbee, Meredith L. Carpenter, Katie Hoefs‐Martin, Samantha A. Brooks, Christa Lafayette, Identification of a novel missense variant in SLC45A2 associated with dilute snowdrop phenotype in Gypsy horses, Animal Genetics, 10.1111/age.12913, 51, 2, (342-343), (2020).
- Julia Metzger, Ottmar Distl, Genetics of Equine Orthopedic Disease, Veterinary Clinics of North America: Equine Practice, 10.1016/j.cveq.2020.03.008, (2020).
- T. Raudsepp, C. J. Finno, R. R. Bellone, J. L. Petersen, Ten years of the horse reference genome: insights into equine biology, domestication and population dynamics in the post‐genome era, Animal Genetics, 10.1111/age.12857, 50, 6, (569-597), (2019).
- H. M. Holl, K. M. Pflug, K. M. Yates, K. Hoefs‐Martin, C. Shepard, D. G. Cook, C. Lafayette, S. A. Brooks, A candidate gene approach identifies variants in 45A2 that explain dilute phenotypes, pearl and sunshine, in compound heterozygote horses, Animal Genetics, 10.1111/age.12790, 50, 3, (271-274), (2019).
- Marina Solé, Michela Ablondi, Amrei Binzer-Panchal, Brandon D. Velie, Nina Hollfelder, Nadine Buys, Bart J. Ducro, Liesbeth François, Steven Janssens, Anouk Schurink, Åsa Viklund, Susanne Eriksson, Anders Isaksson, Hanna Kultima, Sofia Mikko, Gabriella Lindgren, Inter- and intra-breed genome-wide copy number diversity in a large cohort of European equine breeds, BMC Genomics, 10.1186/s12864-019-6141-z, 20, 1, (2019).
- Adam Driver, Frances Henson, Jessica A. Kidd, Luis P. Lamas, Rob Pilsworth, Back Pathology, Equine Neck and Back Pathology, 10.1002/9781118974520, (195-238), (2018).
- Christian J. Posbergh, Geoffrey E. Pollott, Teresa L. Southard, Thomas J. Divers, Samantha A. Brooks, A Nonsynonymous Change in Adhesion G Protein–Coupled Receptor L3 Associated With Risk for Equine Degenerative Myeloencephalopathy in the Caspian Horse, Journal of Equine Veterinary Science, 10.1016/j.jevs.2018.08.010, 70, (96-100), (2018).
- Heather Holl, Ramiro Isaza, Yasmin Mohamoud, Ayeda Ahmed, Faisal Almathen, Cherifi Youcef, Semir Gaouar, Douglas Antczak, Samantha Brooks, A Frameshift Mutation in KIT is Associated with White Spotting in the Arabian Camel, Genes, 10.3390/genes8030102, 8, 3, (102), (2017).
- E. A. Staiger, J. D. Albright, S. A. Brooks, Genome‐wide association mapping of heritable temperament variation in the Tennessee Walking Horse, Genes, Brain and Behavior, 10.1111/gbb.12290, 15, 5, (514-526), (2016).
- Elizabeth A. Staiger, Chia T. Tseng, Donald Miller, Jennifer M. Cassano, Lubna Nasir, Dorian Garrick, Samantha A. Brooks, Douglas F. Antczak, Host genetic influence on papillomavirus‐induced tumors in the horse, International Journal of Cancer, 10.1002/ijc.30120, 139, 4, (784-792), (2016).
- E. A. Staiger, M. A. Abri, C. A. S. Silva, S. A. Brooks, Loci impacting polymorphic gait in the Tennessee Walking Horse1, Journal of Animal Science, 10.2527/jas.2015-9936, 94, 4, (1377-1386), (2016).
- Lauren N. Jacobs, Elizabeth A. Staiger, Julia D. Albright, Samantha A. Brooks, The MC1R and ASIP Coat Color Loci May Impact Behavior in the Horse , Journal of Heredity, 10.1093/jhered/esw007, 107, 3, (214-219), (2016).
- E. A. Staiger, M. A. Al Abri, K. M. Pflug, S. E. Kalla, D. M. Ainsworth, D. Miller, T. Raudsepp, N. B. Sutter, S. A. Brooks, Skeletal variation in Tennessee Walking Horses maps to the LCORL/NCAPG gene region , Physiological Genomics, 10.1152/physiolgenomics.00100.2015, 48, 5, (325-335), (2016).
- H. M. Holl, S. A. Brooks, S. Archer, K. Brown, J. Malvick, M. C. T. Penedo, R. R. Bellone, Variant in the 3 gene associated with 1, a modifier of leopard complex spotting, Animal Genetics, 10.1111/age.12375, 47, 1, (91-101), (2015).
- Molly McCue, Jim Mickelson, Genomic Tools and Resources: Development and Applications of an Equine SNP Genotyping Array, Equine Genomics, 10.1002/9781118522158, (113-124), (2013).
- Molly E. McCue, Danika L. Bannasch, Jessica L. Petersen, Jessica Gurr, Ernie Bailey, Matthew M. Binns, Ottmar Distl, Gérard Guérin, Telhisa Hasegawa, Emmeline W. Hill, Tosso Leeb, Gabriella Lindgren, M. Cecilia T. Penedo, Knut H. Røed, Oliver A. Ryder, June E. Swinburne, Teruaki Tozaki, Stephanie J. Valberg, Mark Vaudin, Kerstin Lindblad-Toh, Claire M. Wade, James R. Mickelson, A High Density SNP Array for the Domestic Horse and Extant Perissodactyla: Utility for Association Mapping, Genetic Diversity, and Phylogeny Studies, PLoS Genetics, 10.1371/journal.pgen.1002451, 8, 1, (e1002451), (2012).




